
Plot accelerometer data for each day with both nonwear time and physical activity intensity categories
Source:R/plot_data_with_intensity.R
plot_data_with_intensity.RdThis function plots accelerometer data with intensity categories against time for each day of measurement, with the possibility to specify the metric to visualize.
Usage
plot_data_with_intensity(
data,
metric = "axis1",
col_time = "time",
col_nonwear = "non_wearing_count",
col_wear = "wearing_count",
valid_wear_time_start = "00:00:00",
valid_wear_time_end = "23:59:59",
zoom_from = "00:00:00",
zoom_to = "23:59:59"
)Arguments
- data
A dataframe obtained using the
prepare_dataset,mark_wear_time, and then themark_intensityfunctions.- metric
A character value to indicate the name of the variable to be plotted against time.
- col_time
A character value to indicate the name of the variable to plot time data.
- col_nonwear
A character value to indicate the name of the variable used to count nonwear time.
- col_wear
A character value to indicate the name of the variable used to count wear time.
- valid_wear_time_start
A character value with the HH:MM:SS format to set the start of the daily period that will be considered for computing valid wear time.
- valid_wear_time_end
A character value with the HH:MM:SS format to set the end of the daily period that will be considered for computing valid wear time.
- zoom_from
A character value with the HH:MM:SS format to set the start of the daily period to visualize.
- zoom_to
A character value with the HH:MM:SS format to set the end of the daily period to visualize.
Examples
# \donttest{
file <- system.file("extdata", "acc.agd", package = "activAnalyzer")
mydata <- prepare_dataset(data = file)
mydata_with_wear_marks <- mark_wear_time(
dataset = mydata,
TS = "TimeStamp",
to_epoch = 60,
cts = "vm",
frame = 90,
allowanceFrame = 2,
streamFrame = 30
)
#> frame is 90
#> streamFrame is 30
#> allowanceFrame is 2
mydata_with_intensity_marks <- mark_intensity(
data = mydata_with_wear_marks,
col_axis = "vm",
equation = "Sasaki et al. (2011) [Adults]",
sed_cutpoint = 200,
mpa_cutpoint = 2690,
vpa_cutpoint = 6167,
age = 32,
weight = 67,
sex = "male",
)
#> You have computed intensity metrics with the mark_intensity() function using the following inputs:
#> axis = vm
#> sed_cutpoint = 200 counts/min
#> mpa_cutpoint = 2690 counts/min
#> vpa_cutpoint = 6167 counts/min
#> equation = Sasaki et al. (2011) [Adults]
#> age = 32
#> weight = 67
#> sex = male
plot_data_with_intensity(
data = mydata_with_intensity_marks,
metric = "vm",
valid_wear_time_start = "00:00:00",
valid_wear_time_end = "23:59:59",
zoom_from = "02:00:00",
zoom_to = "23:58:00"
)
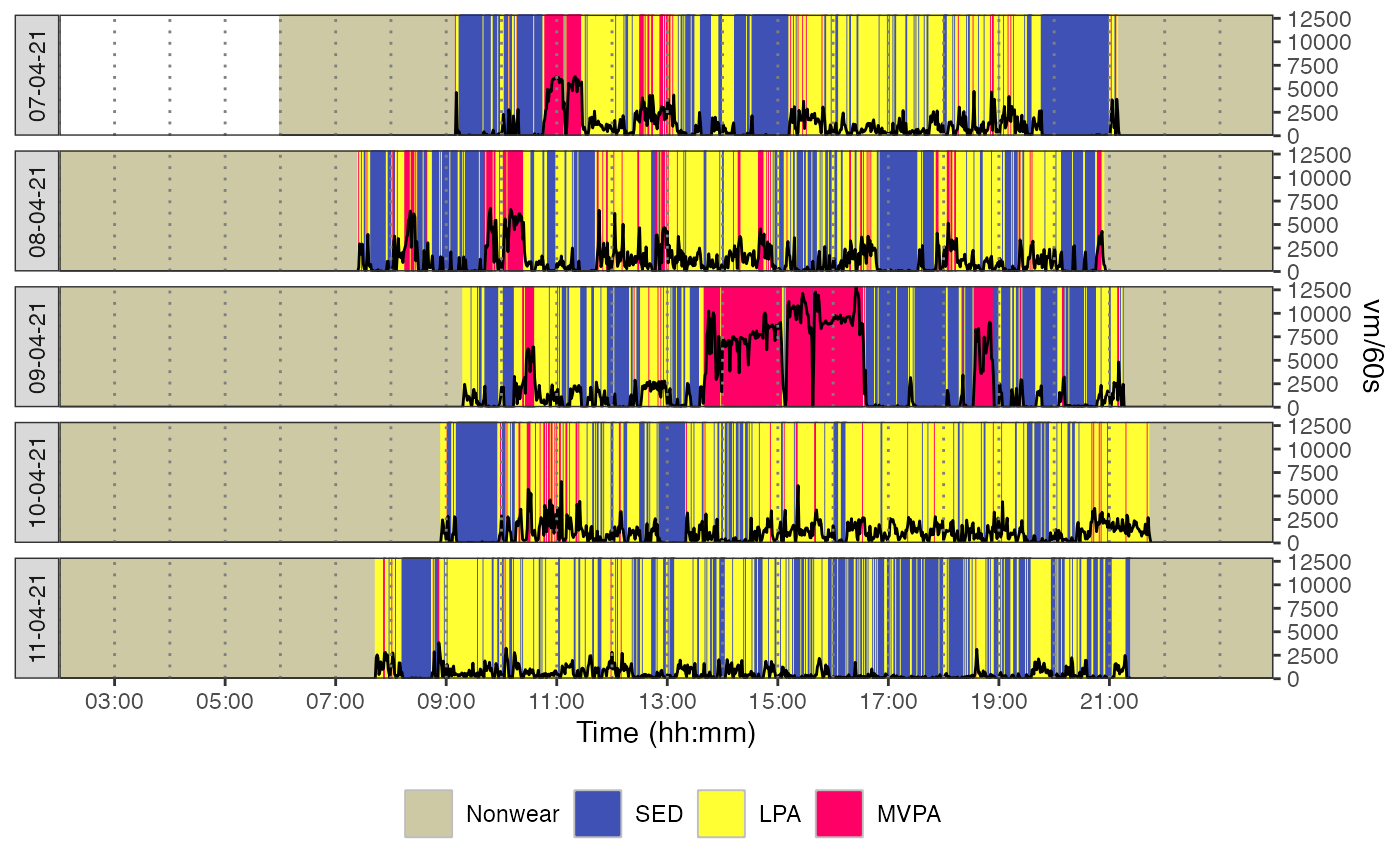 # }
# }