
Create a figure with metrics shown for each day
Source:R/create_fig_res_by_day.R
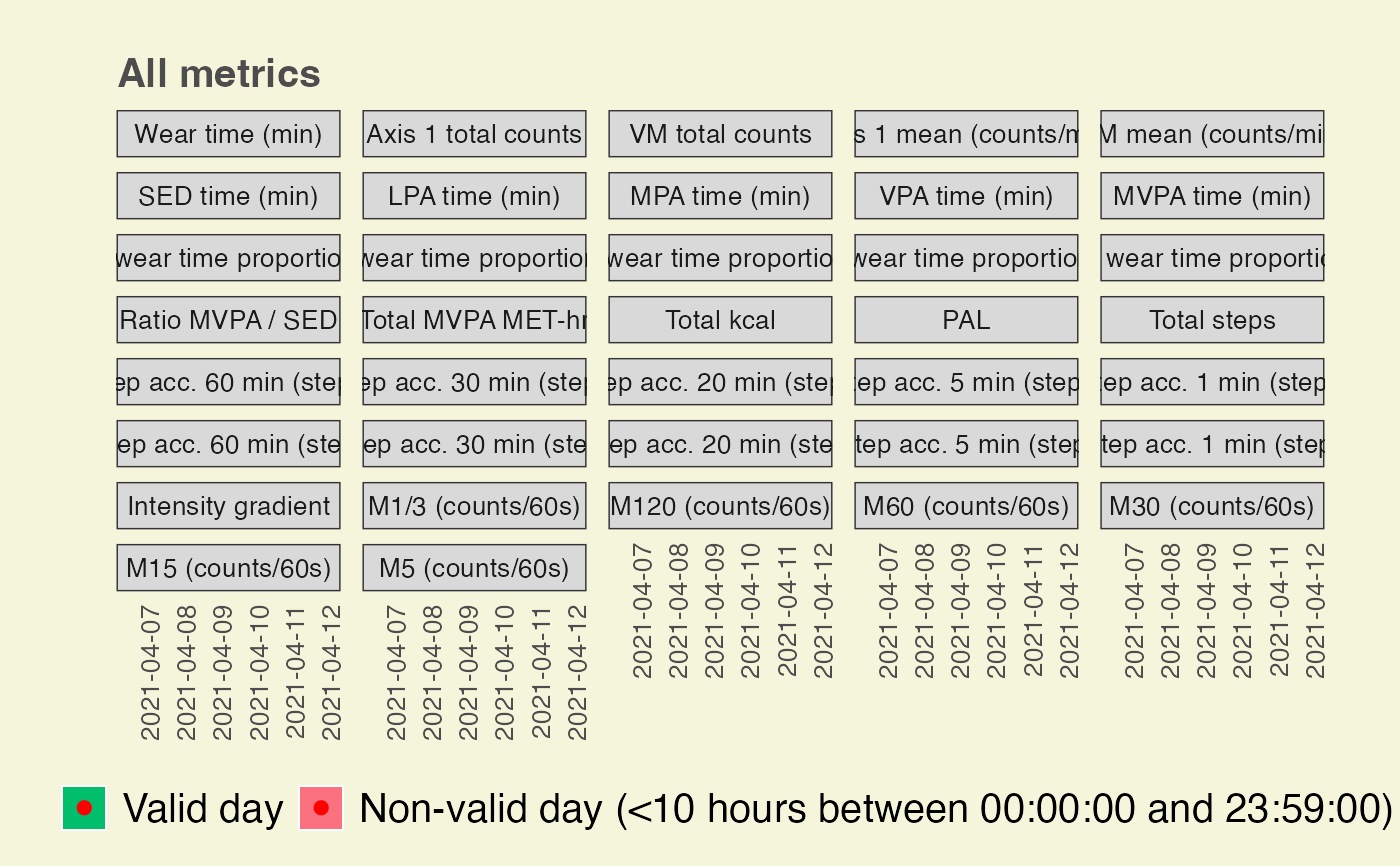
create_fig_res_by_day.RdThe function generates a figure with several common metrics shown for each day of the physical behavior measurement.
Arguments
- data
A dataframe with results obtained using the
prepare_dataset,mark_wear_time,mark_intensity, and then therecap_by_dayfunctions.- minimum_wear_time_for_analysis
A numeric value to indicate the minimum number of hours of wear time that was considered to validate a day.
- start_day_analysis
A character value to indicate the start of the period that was considered to validate a day based on wear time.
- end_day_analysis
A character value to indicate the end of the period that was considered to validate a day based on wear time.
- language
A character value for setting the language with which the figure should be created:
enfor english;frfor french.- metrics
A character value for setting the metrics to be shown in the figure. "volume" refers to "activity volume" metrics, step_acc" refers to "step accumulation" metrics, and "int_distri" refers to intensity distribution metrics. By default, the function provides all computed metrics.
- epoch_label
A character value to be pasted into the names of the variables to build the figure
Examples
# \donttest{
file <- system.file("extdata", "acc.agd", package = "activAnalyzer")
mydata <- prepare_dataset(data = file)
mydata_with_wear_marks <- mark_wear_time(
dataset = mydata,
TS = "TimeStamp",
cts = "vm",
frame = 90,
allowanceFrame = 2,
streamFrame = 30
)
#> frame is 90
#> streamFrame is 30
#> allowanceFrame is 2
mydata_with_intensity_marks <- mark_intensity(
data = mydata_with_wear_marks,
col_axis = "vm",
equation = "Sasaki et al. (2011) [Adults]",
sed_cutpoint = 200,
mpa_cutpoint = 2690,
vpa_cutpoint = 6167,
age = 32,
weight = 67,
sex = "male",
)
#> You have computed intensity metrics with the mark_intensity() function using the following inputs:
#> axis = vm
#> sed_cutpoint = 200 counts/min
#> mpa_cutpoint = 2690 counts/min
#> vpa_cutpoint = 6167 counts/min
#> equation = Sasaki et al. (2011) [Adults]
#> age = 32
#> weight = 67
#> sex = male
summary_by_day <- recap_by_day(
data = mydata_with_intensity_marks,
age = 32,
weight = 67,
sex = "male",
valid_wear_time_start = "07:00:00",
valid_wear_time_end = "22:00:00"
)$df_all_metrics
#> Joining with `by = join_by(date)`
#> Joining with `by = join_by(date)`
#> Joining with `by = join_by(date)`
#> You have computed results with the recap_by_day() function using the following inputs:
#> age = 32
#> weight = 67
#> sex = male
create_fig_res_by_day(summary_by_day,
minimum_wear_time_for_analysis = 10,
start_day_analysis = "00:00:00",
end_day_analysis = "23:59:00",
language = "en")
 # }
# }